A Fully Defined Yeast iVT tRNA Translation Platform and the Synergistic Role of eIF5A and Mg2+
A Fully Defined Yeast iVT tRNA Translation Platform and the Synergistic Role of eIF5A and Mg2+
Jin, Y.
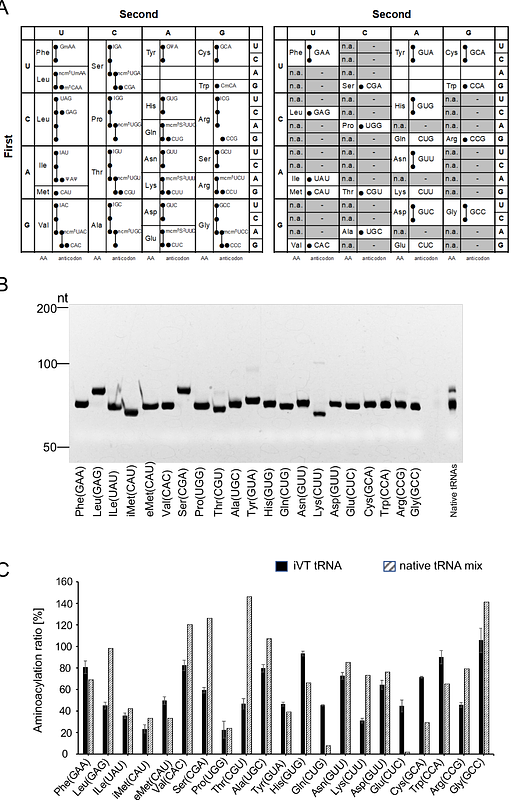
AbstractWe reconstructed Saccharomyces cerevisiae translation with a fully synthetic panel of twenty-one in-vitro-transcribed tRNAs, one isoacceptor per canonical amino acid plus the initiator species. This minimal pool decodes all sixty-one sense codons and, after individual aminoacylation, drives peptide synthesis in a defined yeast PURE system at yields comparable to native tRNAs. Long peptides stall unless supplemented; adding either eukaryotic factor eIF5A or elevated Mg2+ restores activity, and their combination increases nanoluciferase output about fivefold. Alanine scanning shows that basic residues R27 and R87 within the eIF5A core, rather than the hypusine side chain, provide the principal P-site tRNA stabilisation. Higher magnesium alone accelerates elongation but raises UAG and UGA read-through to roughly fifteen percent, revealing a trade-off between speed and fidelity when tRNAs are unmodified. Installing t6A37 or m1G37 on selected synthetic tRNAs further improves processivity. The resulting minimal yet programmable yeast platform enables systematic dissection of modification roles and rapid genetic-code engineering.