Quantification of single cell-type-specific alternative transcript initiation
Quantification of single cell-type-specific alternative transcript initiation
Fu, S.; Wilson, P.; Zhang, B.
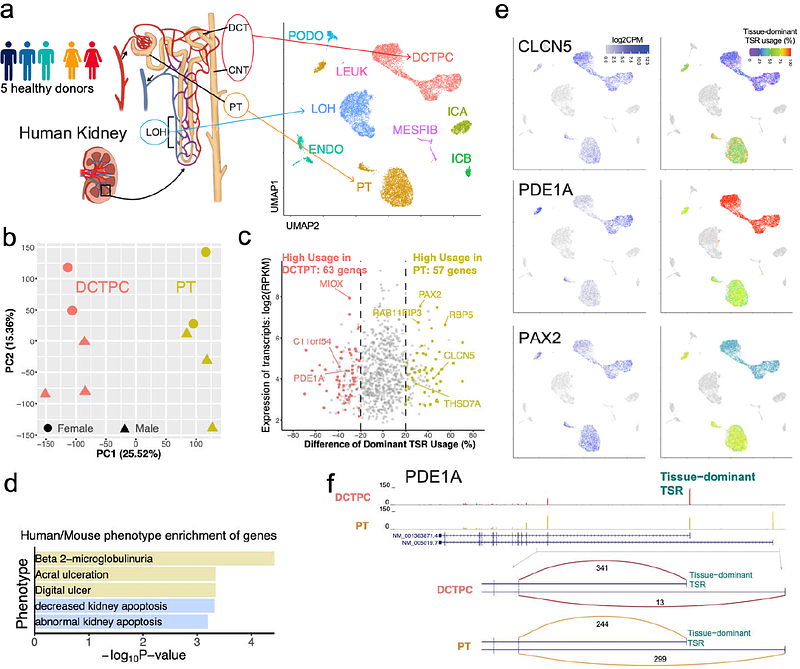
AbstractCells can transcribe different isoforms of a gene by using distinct Transcriptional Start Regions (TSRs), which are recognized by RNA-Polymerase II and regulated by cell-type-specific expressed transcription factors, eventually forming tissue and cell-type-specific expression during development. However, how the distinct TSRs are selectively activated in different tissues and cell types remains largely uncharacterized. To better explore the alternative usage of gene TSRs, we developed TSRdetector, a novel bioinformatic method specifically designed to detect the significant usage alteration of gene TSRs in different tissues, cell types, or diseases. TSRdetector can process either scRNA-seq or bulk-RNA-seq transcriptome data, define dominant TSRs, and compute the differential usage of gene TSRs between given conditions. To demonstrate the capacity of TSRdetector, we applied TSRdetector to analyze a 10X snRNA-seq dataset of healthy and diabetic human kidneys, a Smart-seq2 dataset of mouse B-cell differentiation, and bulk-RNA-seq data of pluripotency transition in human ESCs. In all three analyses, TSRdetectors discovered significant alterations in TSR usage, accompanied by the significant remodeling of the epigenetic landscape. Alteration of TSR usage can change the dominant transcript isoform and further affect the major protein products. Interestingly, a large proportion of these alterations of TSR usage in different cell types did not change the overall gene expression, revealing unique transcription regulations that are independent of expression level. In summary, TSRdetector is a user-friendly package to analyze the differential usage of gene TSRs by using both scRNA-seq and bulk-RNA-seq data, and can be used to explore the alternative transcription initiations of genes at the single cell-type level.