PhysDock: A Physics-Guided All-Atom Diffusion Model for Protein-Ligand Complex Prediction
PhysDock: A Physics-Guided All-Atom Diffusion Model for Protein-Ligand Complex Prediction
Zhang, K.; Ma, Y.; Yu, J.; Luo, H.; Lin, J.; Qin, Y.; Li, X.; Jiang, Q.; Bai, F.; Dou, J.; Zheng, J.; Yu, J.; Sun, L.
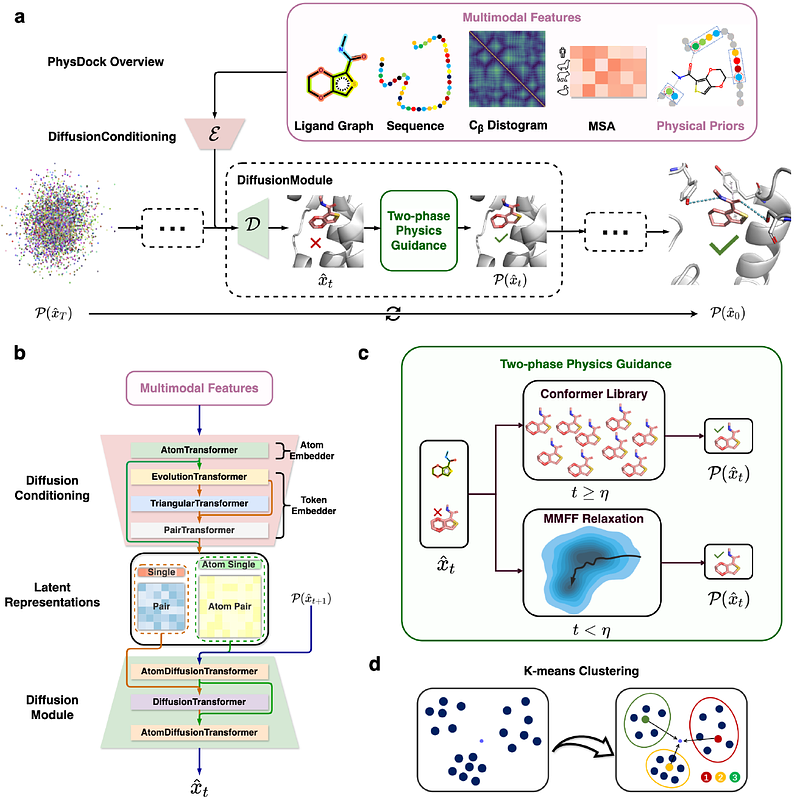
AbstractAccurate prediction of protein-ligand complexes remains a central challenge in structural biology. Traditional methods are computationally inefficient and prone to local minima, whereas deep learning approaches struggle to capture molecular flexibility and physical plausibility. We introduce PhysDock, a physics-guided diffusion model that uniquely integrates (i) all-atom diffusion to model ligand flexibility and protein precision-flexibility; (ii) physical priors as diffusion conditioning, alongside two-phase physics guidance during the denoising diffusion to ensure physical plausibility. PhysDock demonstrates state-of-the-art performance in redocking benchmarks and excels in the more challenging cross-docking assessments involving protein structures resolved with different ligands. For practical utility, PhysDock (i) resolves cannabinoid receptor selectivity across diverse molecules, achieving accuracy comparable to experiments; (ii) effectively distinguishes most drug candidates from weak binders in virtual screening of NTRK3 kinase, while also uncovering novel candidate molecules with structural insights. These results highlight PhysDock as a versatile tool for protein-ligand complex prediction, with substantial potential to accelerate structure-based drug discovery.