PyCycleBio: modelling non-sinusoidal-oscillator systems in temporal biology
PyCycleBio: modelling non-sinusoidal-oscillator systems in temporal biology
Bennett, A. R.; Birchenough, G.; Bojar, D.
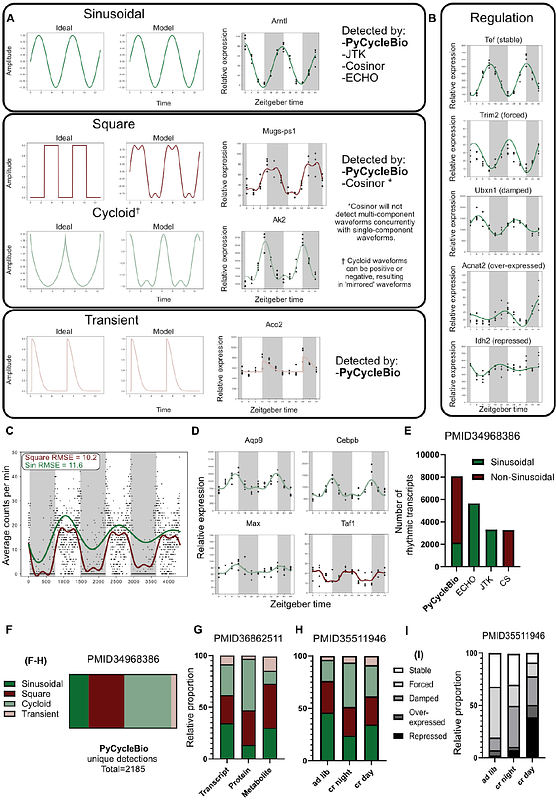
AbstractProtein, mRNA, and metabolite abundances can exhibit rhythmic dynamics, such as during the day/night cycle. Leading bioinformatics platforms for identifying biological rhythms often utilise single-component models of the harmonic oscillator equation, or multi-component models based upon the Cosinor framework. These approaches offer distinct advantages: modelling either temporally-resolved regulatory behaviour via the extended harmonic oscillator equation, or complex rhythmic patterns in the case of Cosinor. Here, we have developed a new platform to combine the advantages of these two approaches. PyCycleBio utilises bounded-multi-component models and modulus operators alongside the harmonic oscillator equation, to model a diverse and interpretable array of rhythmic behaviours, including the regulation of temporal dynamics via amplitude coefficients. We demonstrate increased sensitivity and functionality of PyCycleBio compared to other analytical frameworks, and uncover new relationships between data modalities or sampling conditions with the qualities of rhythmic behaviours from biological datasets- including transcriptomics, proteomics, and metabolomics. We envision that this new approach for disentangling complicated temporal regulation of biomolecules will advance chronobiology and our understanding of physiology. PyCycleBio is available at: https://github.com/Glycocalex/PyCycleBio, and the Python package is available to install at: https://pypi.org/project/pycyclebio/. PyCycleBio can also be used at https://colab.research.google.com/github/Glycocalex/PyCycleBio/blob/main/PyCycleBio.ipynb with no installations necessary.