Lazypipe3: Customizable Virome Analysis Pipeline Enabling Fast and Sensitive Virus Discovery from NGS data
Lazypipe3: Customizable Virome Analysis Pipeline Enabling Fast and Sensitive Virus Discovery from NGS data
Weinstein, I.; Vapalahti, O.; Kant, R.; Smura, T.
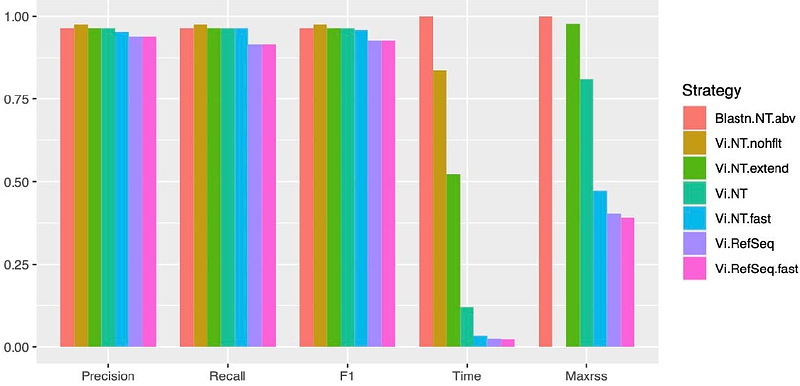
AbstractMetagenomic next-generation sequencing (mNGS) has revolutionized virus detection across clinical, animal, and wildlife samples, enabling the discovery of both known and novel viruses. However, efficient identification of viromes from mNGS data depends heavily on bioinformatics pipelines. To contribute to publicly available pipelines, we introduce here Lazypipe3 with multiple new features including flexible annotation strategies that can be adjusted to match different datasets and research goals. Using a synthetic benchmark, we show that a simple two-round annotation strategy designed for speed and low false positive rate required in virus diagnostics, can reduce execution time to a fraction of BLAST search without any loss in accuracy. Additionally, using real data from mosquito samples we show that annotation strategies based on combinations of fast-to-accurate homology searches reduce execution time from 5 to 20-fold compared to BLASTN/BLASTP baseline. Using results from one of these \"annotation chains\" we were able to discover and characterise multiple novel whole-genome viral sequences that were missed by previous Lazypipe1 and BLASTX analysis. In Lazypipe3 we also implemented and benchmarked an improved background filtering and novel interactive reports for visualising coverage and placement of contigs relative to reference genomes. Lazypipe3 represents a highly efficient and versatile tool for virome analysis, offering customizable and transparent workflows that can facilitate virus discovery and diagnostics in diverse mNGS applications.