multideconv - Integrative pipeline for cell type deconvolution from bulk RNAseq using first and second generation methods
multideconv - Integrative pipeline for cell type deconvolution from bulk RNAseq using first and second generation methods
Hurtado, M.; Essabbar, A.; Khajavi, L.; Pancaldi, V.
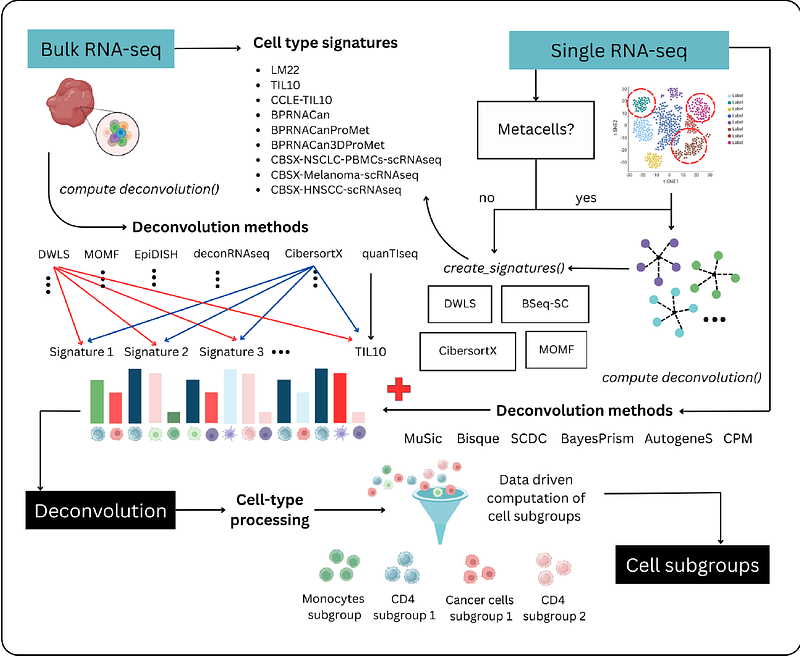
AbstractSummary: Computational methods for cell type deconvolution from bulk RNA-seq data have been increasing in the last years, but their high feature complexity and variability across methods and signatures limit their utility and effectiveness for patient stratification. Applying multiple combinations of deconvolution methods and signatures often results in hundreds of redundant or contradictory cell features describing the tumor microenvironment (TME). Benchmarking efforts of deconvolution methods to look for the best one are inherently limited by the lack of bias-free ground truth, often yielding inconsistent results or no consensus. To address this, we present multideconv, an R package that reduces dimensionality and eliminates redundancy through unsupervised filtering and iterative correlation analyses. Built on top of existing frameworks like immunedeconv (Sturm et al. 2019) and omnideconv (Dietrich et al. 2024), multideconv harmonizes outputs across methods to identify robust cell subgroups and mitigate signature-driven heterogeneity.