SynVerse: A Framework for Systematic Evaluation of Deep Learning Based Drug Synergy Prediction Models
SynVerse: A Framework for Systematic Evaluation of Deep Learning Based Drug Synergy Prediction Models
TASNINA, N.; Haghani, M.; Murali, T. M.
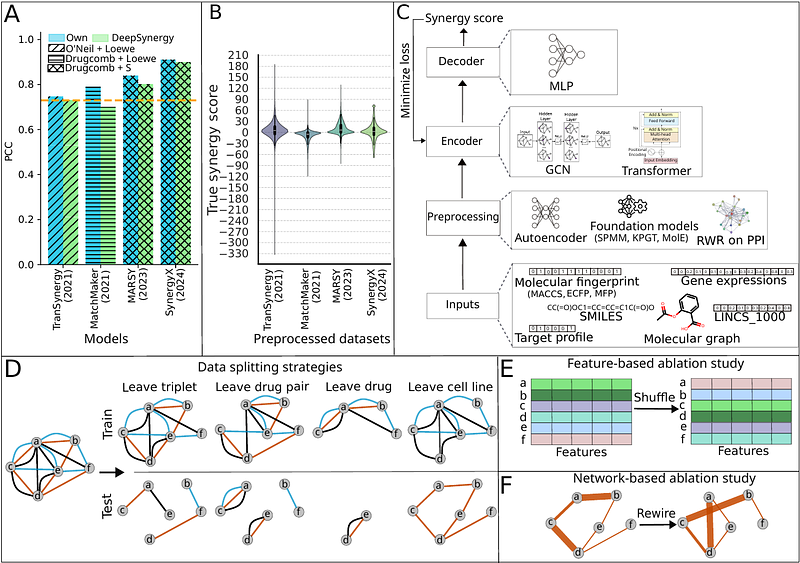
AbstractSynergistic drug combinations are often used to treat cancer. Experimental exploration of all possibilities is expensive. Deep learning (DL) for predicting the synergy of drug pairs in specific cell lines might provide an alternative. However, current approaches often suffer from data leakage. They also lack systematic ablation studies. To address these gaps, we propose SynVerse, a comprehensive evaluation framework featuring four data-splitting strategies to assess DL model generalizability and three ablation studies: module-based, feature shuffling, and a novel network-based approach to disentangle factors influencing model performance. We evaluated sixteen models incorporating eight drug- and cell line-specific features, five preprocessing techniques, and two widely used encoders. Our analysis revealed several insights. None of the models outperformed a naive baseline using one-hot encoding as features. Biologically meaningful drug or cell line features and drug-drug interactions were not the drivers of predictive performance. All models demonstrated poor generalization to unseen drugs and cell lines. SynVerse emphasizes the need for substantial improvements before computational predictors can reliably support experimental and clinical settings.