Pathogenic Missense Mutations in Intrinsically Disordered Regions Reveal Functional and Clinical Signals
Pathogenic Missense Mutations in Intrinsically Disordered Regions Reveal Functional and Clinical Signals
Deutsch, N.; Erdos, G.; Dosztanyi, Z.
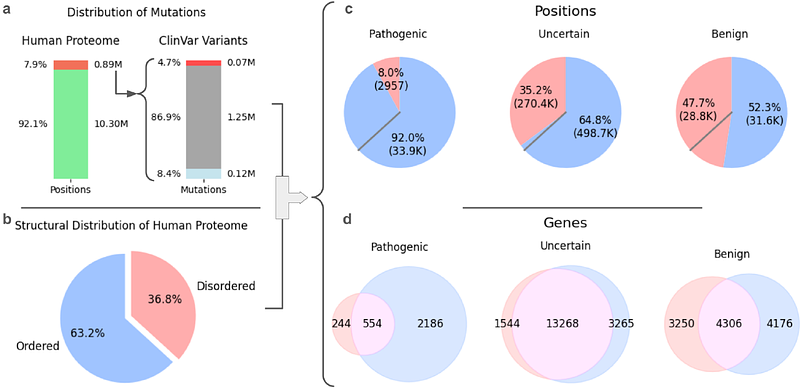
AbstractIntrinsically disordered regions (IDRs) play essential roles in signaling and regulation but remain under-characterized in the context of human disease. We analyzed nearly one million ClinVar missense variants, focusing on the subset within IDRs defined by curated and predicted annotations. Pathogenic variants were significantly enriched in short linear motifs (SLiMs) and disordered binding regions, consistent with their functional importance. To extend these insights beyond existing annotations, we applied AlphaMissense, a deep learning model for variant effect prediction, revealing localized \'island-like\' pathogenicity patterns within IDRs. Based on these signals, we developed a classifier to prioritize predicted ELM motifs (PEMs), identifying motifs linked to pathogenicity and major disease classes, including neurological, cardiovascular, and cancer-associated genes. Case studies in POLK, FOXP2, and LMOD3 demonstrate how this framework yields mechanistic insight into disease-relevant mutations within disordered protein regions, advancing clinical interpretation in the disordered proteome.