Improving RNA Secondary Structure Prediction Through ExpandedTraining Data
Improving RNA Secondary Structure Prediction Through ExpandedTraining Data
Langeberg, C. J.; Kim, T.; Nagle, R.; Meredith, C.; Garuadapuri, D. A.; Doudna, J. A.; Cate, J. H. D.
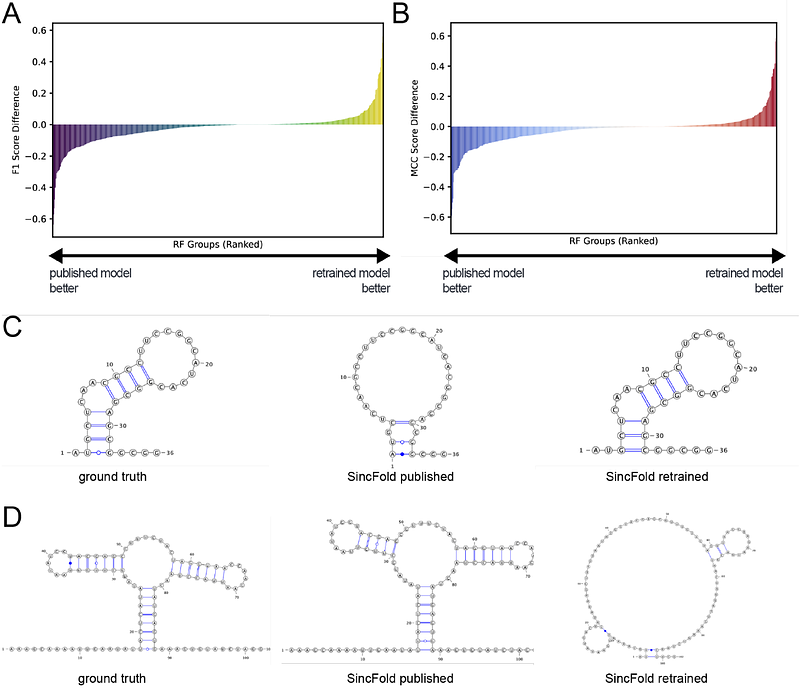
AbstractIn recent years, deep learning has revolutionized protein structure prediction, achieving remarkable speed and accuracy. RNA structure prediction, however, has lagged behind. Although several methods have shown moderate success in predicting RNA secondary and tertiary structures, none have reached the accuracy observed with contemporary protein models. The lack of success of these RNA structure prediction models has been proposed to be due to limited high-quality structural information that can be used as training data. To probe this proposed limitation, we developed a large and diverse dataset comprising paired RNA sequences and their corresponding secondary structures. We assess the utility of this enhanced dataset by retraining two deep learning models, SincFold and MXfold2. We find that SincFold exhibited improved generalization to some previously unseen RNA families, enhancing its capability to predict accurate de novo RNA secondary structures. By contrast, retraining MXfold2 proved too computationally expensive for the large RNASSTR dataset and did not achieve high performance on the testing set. The RNASSTR dataset provides a substantial advance for RNA structure modeling, laying a strong foundation for the development of future RNA secondary structure prediction algorithms.