PEtab.jl: Advancing the Efficiency and Utility of Dynamic Modelling
PEtab.jl: Advancing the Efficiency and Utility of Dynamic Modelling
Cvijovic, M.; Persson, S.; Frohlich, F.; Grein, S.; Lomna, T.; Ognissanti, D.; Hassselgren, V.; Hasenauer, J.
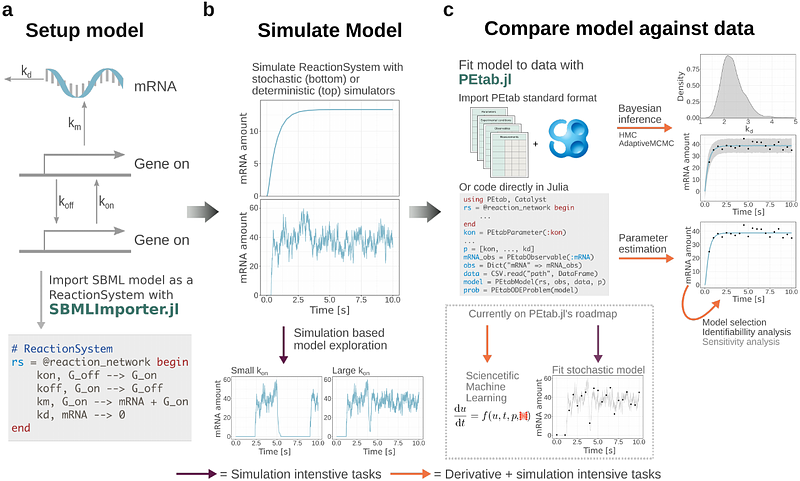
AbstractDynamic models are useful to study processes ranging from cell signalling to cell differentiation. Common modelling workflows, such as model exploration and parameter estimation, are computationally demanding. The Julia programming language is a promising tool to address these computational challenges. To evaluate it, we developed SBMLImporter.jl and PEtab.jl, a package for model fitting. SBMLImporter.jl was used to evaluate different stochastic simulators against PySB and RoadRunner, overall Julia simulators proved fastest. For Ordinary Differential Equations (ODE) models solvers, gradient methods, and parameter estimation performance were evaluated using PEtab benchmark problems. For the latter two tasks PEtab.jl was compared against pyPESTO, which employs the high-performance AMICI library. Guidelines for choosing ODE solver were produced by evaluating 31 ODE solvers for 29 models. Further, by leveraging automatic differentiation PEtab.jl proved efficient and, for up to medium-sized models, was often at least twice faster than pyPESTO, showcasing how Julia\'s ecosystem can accelerate modelling workflows.