Explainable deep learning for identifying cancer driver genes based on the Cancer Dependency Map
Explainable deep learning for identifying cancer driver genes based on the Cancer Dependency Map
Yin, Q.; Chen, L.
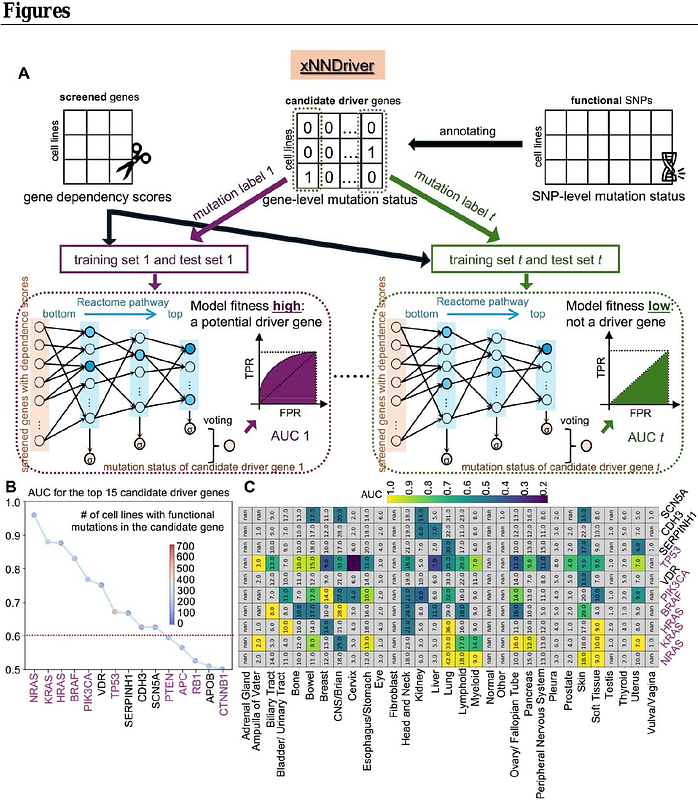
AbstractDetecting cancer driver genes and mutations is critical but challenging in cancer biology, essential for understanding tumor progression and developing targeted therapies. In this study, we leveraged the Cancer Dependency Map (DepMap) to identify potential cancer driver genes and infer multi-driver mutations within tumor samples. We developed xNNDriver, a biologically informed, supervised deep learning model that links the mutation status of a candidate cancer driver gene to genome-wide dependency scores. The model\'s fitness provides a quantifiable measure of a gene\'s likelihood to act as a cancer driver. Our model successfully recovers well-established drivers, including NRAS, KRAS, HRAS, and BRAF, and identifies novel candidates such as VDR. The model\'s interpretability further illuminates downstream pathways regulated by driver genes. To complement the gene-by-gene analysis, we built xAEDriver, an unsupervised explainable autoencoder that infers multiple driver variant representations (DVRs) simultaneously. These DVRs capture the mutation patterns across multiple potential driver variants in tumor samples. Notably, DVR-based stratification of cell lines uncovers distinct drug response profiles, further demonstrating the model\'s translational potential. Overall, our framework offers a robust and interpretable approach to cancer driver gene discovery and mutation pattern analysis, offering new insights into cancer biology.