Genome-Wide Identification of Conditionally Essential Genes Supporting Streptococcus suis Growth in Serum and Cerebrospinal Fluid
Genome-Wide Identification of Conditionally Essential Genes Supporting Streptococcus suis Growth in Serum and Cerebrospinal Fluid
Juanpere Borras, M.; Zhao, T.; Boekhorst, J.; Fernandez-Ciruelos, B.; Sanyal, R.; Arifa, N.; Wagenaar, T.; van Baarlen, P.; Wells, J.
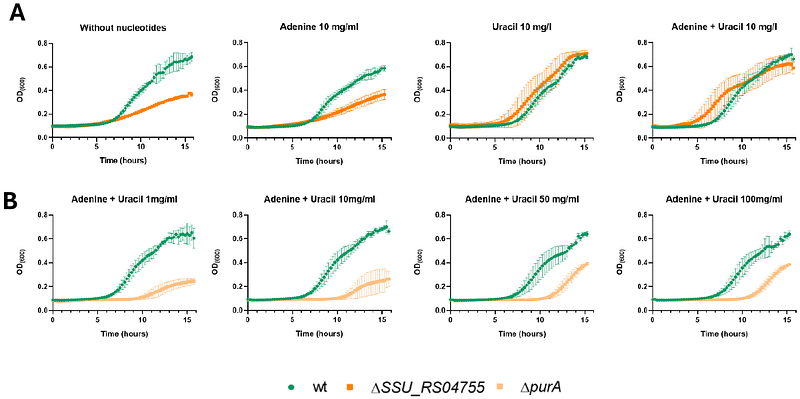
AbstractStreptococcus suis is a major cause of sepsis and meningitis in pigs, and zoonosis through the emergence of disease-associated lineages. The ability of S. suis to adapt and survive in host environments, such as blood and cerebrospinal fluid (CSF), is important for pathogenesis. Here, we used Tn-Seq coupled with Nanopore sequencing to identify conditionally essential genes for growth of S. suis P1/7 in active porcine serum (APS) and CSF derived from choroid plexus organoids (Zhao et al., 2025). Through comparative fitness analyses we identified 33 conditionally essential genes (CEGs) supporting growth in APS and 25 CEGs in CSF, highlighting the importance of pathways involved in amino acid transport, nucleotide metabolism, and cell envelope integrity. Notably, the LiaFSR regulatory system and multiple ABC transporters were important for proliferation. We also identified several genes of unknown function as essential for growth, pointing to previously unrecognized genetic factors involved in S. suis adaptation during infection. These findings provide new insights into the genetic requirements for S. suis survival in host-like environments and a deeper understanding of its ability to adapt to distinct physiological niches.