Top-Down Scoring of Spectral Fitness by Image Analysis for Protein Structure Validation
Top-Down Scoring of Spectral Fitness by Image Analysis for Protein Structure Validation
Harding, B. D.; DeZonia, B.; Garg, R.; Hu, Z.; Delaglio, F.; Grant, T.; Rienstra, C. M.
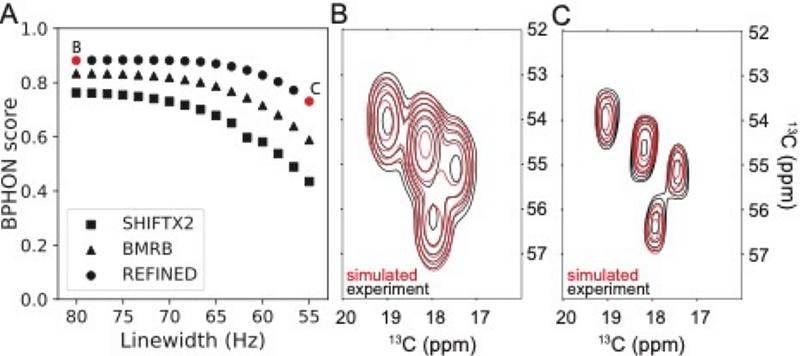
AbstractNuclear magnetic resonance (NMR) spectroscopy is a powerful technique for protein structure determination, but traditional approaches require extensive manual assignment of hundreds to thousands of resonances. Here we present NMRFAM-BPHON, a novel \"top-down\" approach that treats experimental NMR spectra as continuous grayscale images and quantitatively scores the agreement with simulated spectra generated from candidate protein structures. This method does not require complete resonance assignments, though it can incorporate experimental chemical shifts when available to improve performance. The simulated spectra are generated from postulated resonance assignments, which can be derived either from empirical database predictions, direct interpretation, or a hybrid combination. BPHON employs a physics-based approximate polarization transfer model to predict cross-peak intensities from the internuclear distances in the decoy structure, and models the peak lineshapes using empirical, bulk T2 relaxation rates and literature values for scalar couplings. The resulting simulated spectra are scored relative to the experimental data by normalized cross correlation, yielding a fitness score between 0 and 1. We demonstrate BPHON\'s ability to discriminate structural models, particularly in the case of 13C-detected magic angle spinning solid-state NMR spectra. The software is packaged with a user-friendly graphical user interface for ChimeraX, enabling advanced NMR analysis accessible without requiring extensive manual analysis.