Sperrfy the brain: A data-driven realization of Sperry's Chemoaffinity theory in the neural connectome
Sperrfy the brain: A data-driven realization of Sperry's Chemoaffinity theory in the neural connectome
Koike, J.; Yada, Y.; Hira, R.; Naoki, H.
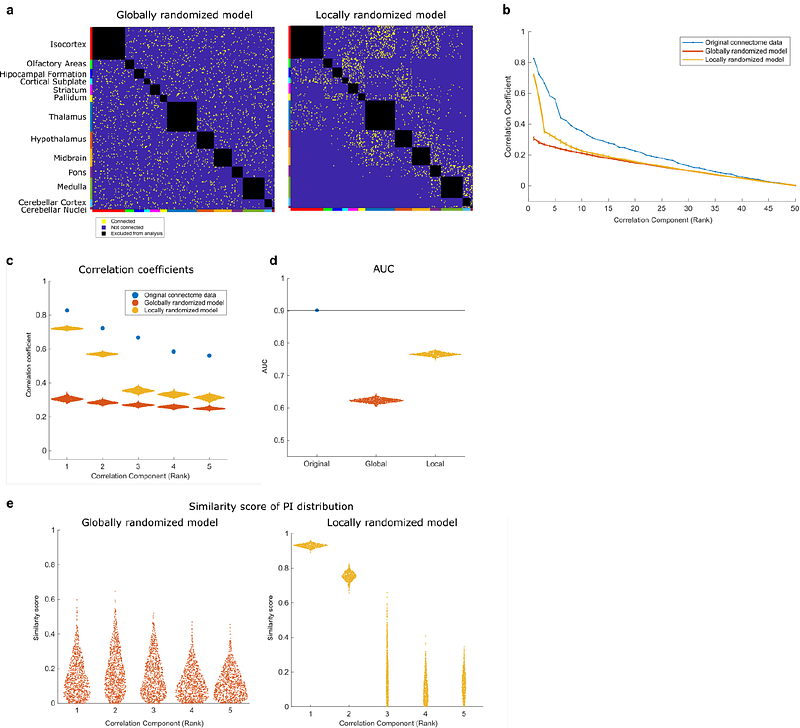
AbstractUnderstanding how brain-wide neural circuits are genetically wired remains a fundamental question in neuroscience. While Sperry\'s chemoaffinity theory posits that molecular gradients provide positional cues for axonal projections, its application has been largely limited to localized sensory systems. Here, we present SPERRFY (Spatial Positional Encoding for Reconstructing Rules of axonal Fiber connectivitY), a data-driven framework that operationalizes Sperry\'s theory at the whole-brain scale. By integrating connectomic data with spatial transcriptomic profiles from the Allen Mouse Brain Atlas, SPERRFY deciphers latent positional gradients that underlie axonal wiring. Using canonical correlation analysis (CCA), we extract top gradient pairs that align with observed neural connectivity patterns, capturing both global (inter-regional) and local (intra-regional) organizational principles. Connectivity reconstruction based on these gradients achieves high predictive performance (AUC = 0.90), and permutation-based null models confirm the biological relevance of the inferred structures. Furthermore, SPERRFY identifies candidate genes that may contribute to positional wiring information, providing molecular insight into the developmental logic of brain-wide circuitry. Our results extend Sperry\'s foundational theory beyond the sensory domain, offering a unified, data-driven framework for understanding genetically encoded connectivity across the entire brain.