G34R cancer mutation alters the conformational ensemble and dynamics of the histone H3.3 tails
G34R cancer mutation alters the conformational ensemble and dynamics of the histone H3.3 tails
Fuchs, H. A.; Peng, Y.; Ayyappan, S.; Zhang, H.; Rosas, R.; Panchenko, A.; Musselman, C. A.
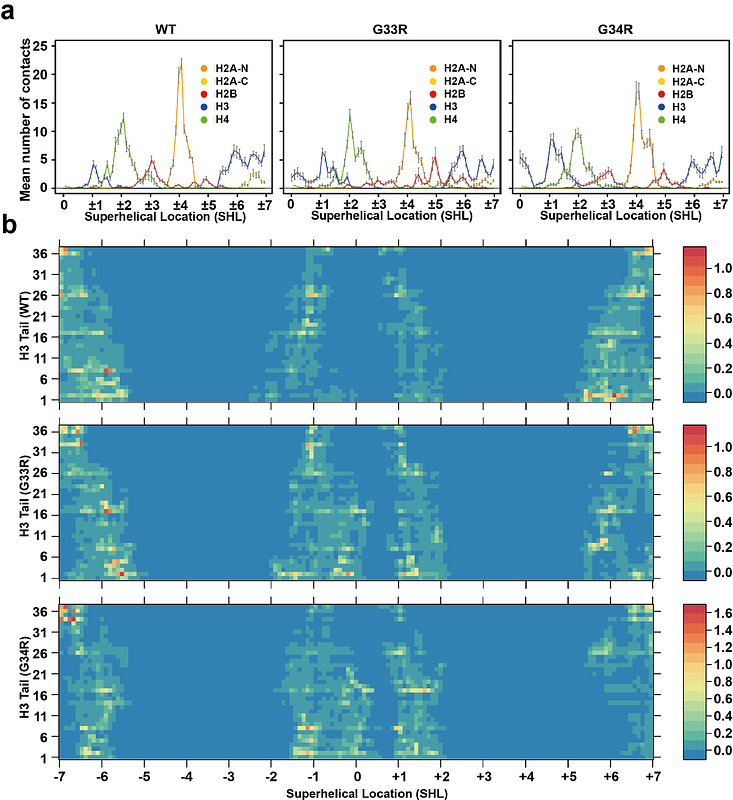
AbstractIn recent years, mutations in the histone variant H3.3 have been discovered in pediatric and adult gliomas and osteosarcomas. One of these is the G34R mutation in the H3.3 N-terminal tail. While this mutation is known to disrupt epigenomic pathways, the effects on nucleosome structure itself have not been explored. In light of recent studies, that have demonstrated that the interaction of the H3 tail with nucleosomal and linker DNA is driven in large part by arginine residues, we sought to determine if the G34R cancer mutation and adjacent G33R mutation, not observed in cancer, directly alter nucleosome structural dynamics. Using NMR spectroscopy and molecular dynamics simulations, we investigate the effects of these mutations on the H3 tail in the context of the nucleosome. Our results show that both of these mutations enhance the association of the H3 tails with DNA and decrease the conformational dynamics around the site of mutation. Moreover, our results reveal that both mutations lead to changes in the conformational ensemble of the entire tail, re-positioning it on the nucleosomal DNA as well as promoting intra-tail interactions. This has implications for the accessibility of the H3 tail to effector proteins and for changes in higher-order chromatin structure resulting from a shift in intra- versus inter-nucleosome contacts mediated by the H3 tail.