Mechanistic models of asymmetric hand-over-hand translocation and nucleosome navigation by CMG helicase
Mechanistic models of asymmetric hand-over-hand translocation and nucleosome navigation by CMG helicase
Nagae, F.; Murata, Y.; Yamauchi, M.; Takada, S.; Terakawa, T.
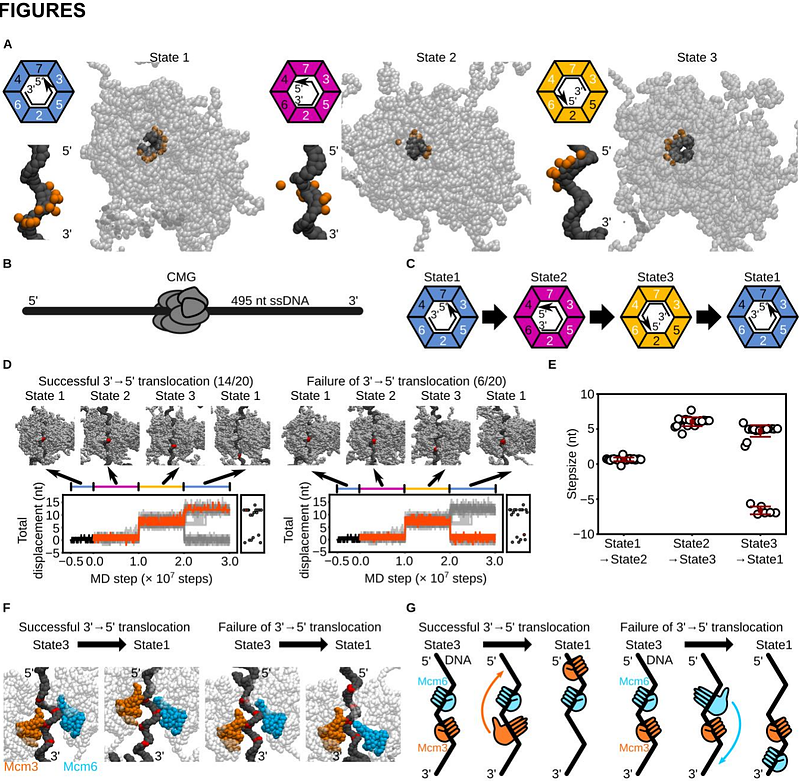
AbstractFaithful replication of eukaryotic chromatin requires the CMG helicase to translocate directionally along single-stranded DNA (ssDNA) while unwinding double-stranded DNA (dsDNA) and navigating nucleosomes. However, the mechanism by which CMG achieves processive translocation and deals with nucleosomal barriers remains incompletely understood. Here, using coarse-grained molecular dynamics simulations with ATP-driven conformational switching, we showed that asymmetric rotational transitions among four distinct ssDNA-binding states enable CMG to achieve directional translocation and DNA unwinding. We further demonstrated that the fork protection complex (Csm3/Tof1) and RPA enhance processivity through distinct mechanisms: Csm3/Tof1 grips the parental duplex to suppress backtracking, while RPA alleviates lagging-strand clogging. Upon nucleosome encounter, Csm3/Tof1 promoted partial unwrapping of the entry DNA, but further progression was energetically restricted near the nucleosomal dyad. The histone chaperone FACT lowered this barrier and simultaneously prevented inappropriate histone transfer to the lagging strand. Our results provide mechanistic insights into how the eukaryotic replisome coordinates helicase activity, nucleosome navigation, histone chaperon function, and histone recycling during eukaryotic DNA replication.