Vernalization regulatory network identifies potential novel functions for ZCCT genes within HvVRN2
Vernalization regulatory network identifies potential novel functions for ZCCT genes within HvVRN2
Montardit-Tarda, F.; Puyo, I.; Contreras-Moreira, B.; Karsai, I.; Borrill, P.; Casas, A. M.; Igartua, E.
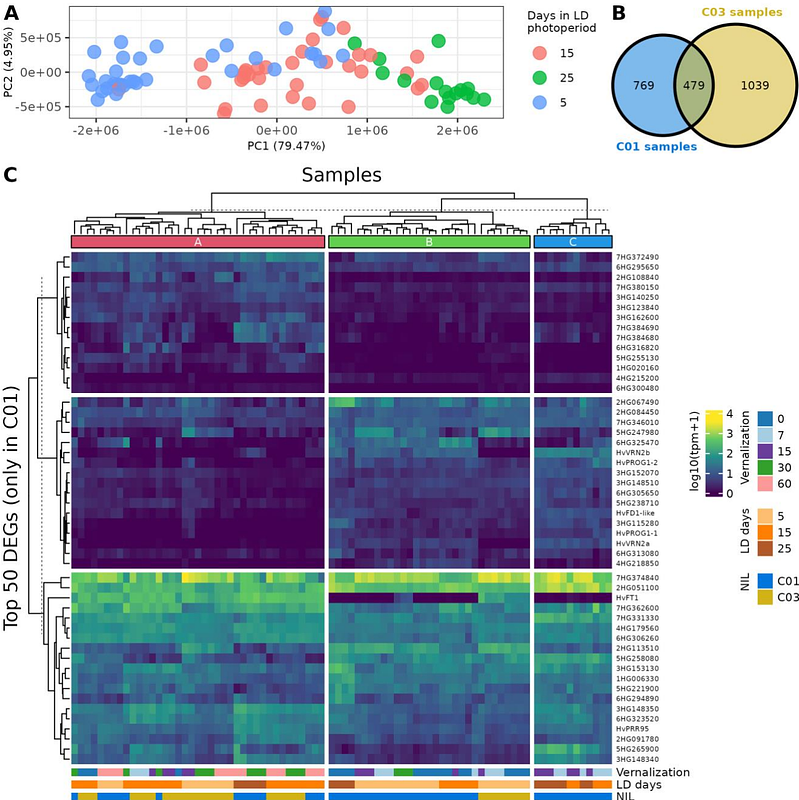
AbstractFlowering is a key yield determinant, controlled by environmental and genetic factors. Vernalization requirement, a prolonged cold period to promote flowering, is determined in barley (Hordeum vulgare L.) by the allele in HvVRN1 and the presence of locus HvVRN2, which consists of two ZCCT genes: HvVRN2a and HvVRN2b. While the effect of HvVRN2 presence is well-known, its outcome on the transcriptome and other genes outside the flowering/vernalization pathway remains unknown. Near-isogenic lines with presence/absence of HvVRN2, were subjected to different lengths of vernalization treatments, identifying the expression dynamics of both HvVRN2 genes and wider transcriptional responses controlled by VRN2. Our work revealed previously unknown differences in expression levels of the two HvVRN2 genes, in responses to its main repressor, HvVRN1, and an effect on tillering. Besides, we shed new light on the repressing mechanism of VRN2 on flowering, using protein modelling. A regulatory network analysis pointed at new candidate genes for flowering regulation related to the vernalization pathway, and suggested additional biological roles for HvVRN2b. This study re-establishes HvVRN2 as a central gene for understanding barley responses to environmental cues, beyond its previously accepted simple role, and expands the catalogue of genes related to the vernalization pathway, which may be targeted in barley breeding.