Bacterial metabolic signatures in MASLD predicted through gene-centric studies in stool metagenomes
Bacterial metabolic signatures in MASLD predicted through gene-centric studies in stool metagenomes
Medina, J. M.; Iruzubieta, P.; Fernandez-Lopez, R.; Crespo, J.; de la Cruz, F.
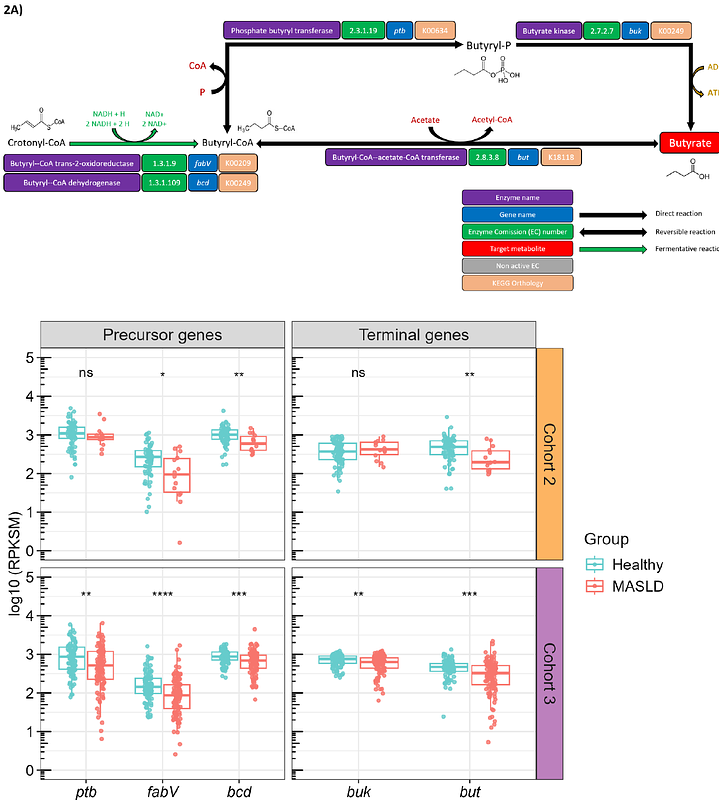
AbstractMetabolic dysfunction-associated steatotic liver disease (MASLD) is a multifactorial condition influenced by the gut microbiome (GM). While previous studies have reported inconsistent associations between MASLD and key microbial clades using low-resolution 16S rRNA profiling, we employed high-resolution metagenomic sequencing and multi-marker taxonomic classification across three independent cohorts to identify robust microbial and functional signatures of MASLD. We consistently detected a depletion of Agathobacter rectalis, a known butyrate producer, in MASLD patients. Functionally, MASLD was characterized by a depletion of genes involved in butyrate and methane biosynthesis -particularly within the crotonyl-butyryl-CoA axis- alongside an enrichment of genes driving the production of endogenous alcohols such as ethanol and 1-propanol. Genes encoding these fermentative pathways, often organized in operons like pdu and tor, were more abundant in MASLD samples, indicating a potential shift toward alcohol-producing metabolism. These geno-metabolic changes were accompanied by a broader displacement of beneficial taxa and an increase in accessory gene content across the GM, underscoring the limitations of taxonomy-based disease associations. Many of the differentially abundant genes were also found on plasmids, suggesting that horizontal gene transfer contributes to strain-level metabolic variability relevant to MASLD progression. Our findings support a model in which GM-driven metabolic shifts -rather than taxonomic changes alone- play a central role in MASLD pathogenesis, highlighting the importance of functional and mobile genetic element profiling for uncovering mechanistic links between the microbiome and liver disease.