Contributions of Folded and Disordered Domains to RNA Binding by HNRNPR
Contributions of Folded and Disordered Domains to RNA Binding by HNRNPR
Guzman, B. B.; Goda, G. A.; Jimenez, A.; Martyr, J. G.; Hu, Y.; Cavazos, F. F.; Aleman, M. M.; Dominguez, D.
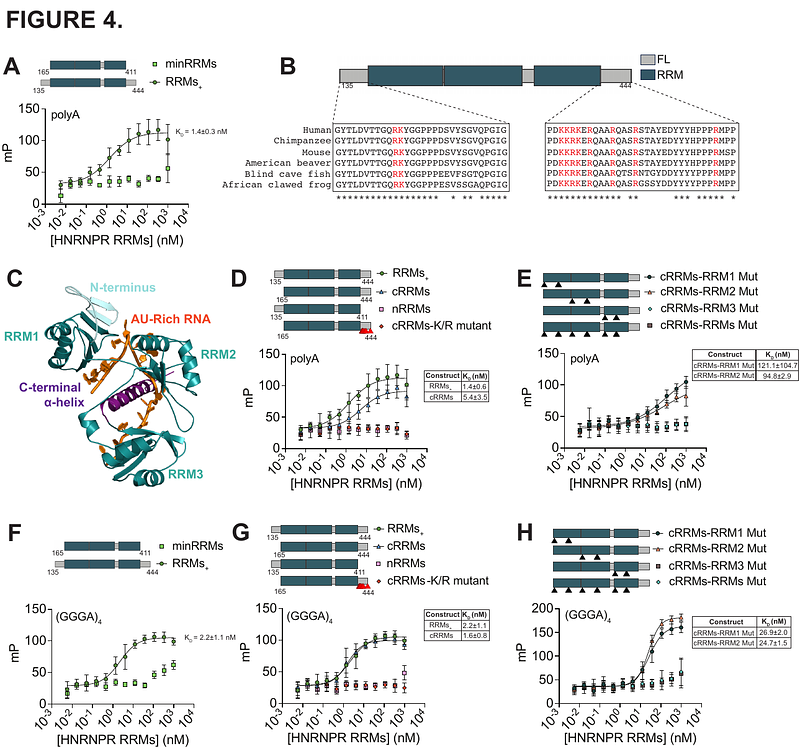
AbstractRNA binding proteins (RBPs) interact with and tightly regulate the fate of messenger RNAs but how RNA targets are recognized remains a challenging question. RBPs often contain multiple domains known to directly bind RNA, such as RNA recognition motifs (RRMs), as well as domains whose RNA binding capacity remains incompletely understood, e.g., low complexity domains (LCDs). Here, we dissect HNRNPR, an RBP with three RRMs and an arginine-glycine rich (RG-rich) LCD. We apply unbiased high-throughput biochemical approaches and identify critical RNA binding domains that confer specificity. We show that not all RRMs contribute equally to binding and find that RRM3, along with a downstream C-terminal charged region, are required for RNA binding. We find that HNRNPR also binds RNA G-quadruplexes (rG4s) and map multiple rG4 binding sites including RRM3 with the C-terminal charged region and RG-rich regions within the LCD. We dissect rG4 specificity for the full length HNRNPR and LCD using a newly created RNA pool focused on rG4s and reveal that binding is dependent on RNA folding and find specific rG4 features that enhance HNRNPR-rG4 interactions. Our work highlights the complexity of RBP-RNA interactions and motivates the study of disordered regions as RNA binding domains.