CRISPRi perturbation screens and eQTLs provide complementary and distinct insights into GWAS target genes
CRISPRi perturbation screens and eQTLs provide complementary and distinct insights into GWAS target genes
Ghatan, S.; Panten, J.; Oliveros, W.; Sanjana, N. E.; Morris, J. A.; Lappalainen, T.
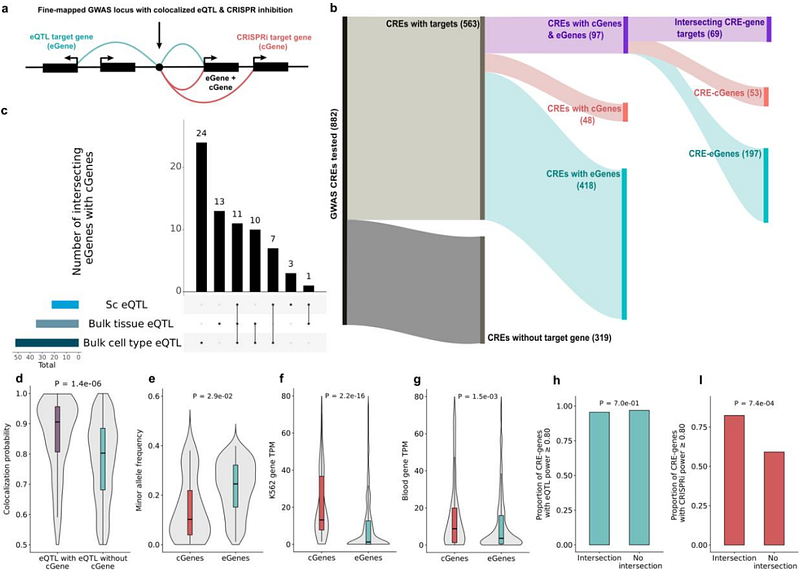
AbstractMost genetic variants associated with human traits and diseases lie in noncoding regions of the genome 1, and a key challenge is to determine which genes they affect 2,3. A common approach has been to leverage associations between natural genetic variation and gene expression to identify expression quantitative trait loci (eQTLs) in the population 4,5, while a newer method uses pooled CRISPR interference (CRISPRi) perturbations of noncoding loci with single-cell transcriptome sequencing 6,7. Here, we systematically compared the results from these approaches across hundreds of genomic regions associated with blood cell traits. We find that while the two approaches often identify the same target genes, they also capture distinct features of gene regulation. CRISPRi tends to detect genes that are physically closer to regulatory variants and more constrained, whereas eQTL studies are sensitive to detecting multiple, often distal, genes. Comparing these discoveries to a gold-standard set of genes linked to blood traits, the two approaches provide highly complementary insights with distinct strengths and caveats. Our results provide insights for improved design of CRISPRi and eQTL studies and indicate the potential for a powerful toolkit for interpretation of disease-associated loci.