Alpha-Synuclein Fibril Structures Cluster into Distinct Classes
Alpha-Synuclein Fibril Structures Cluster into Distinct Classes
Milchberg, M. H.; Warmuth, O. A.; Borcik, C. G.; Dhavale, D. D.; Wright, E. R.; Kotzbauer, P. T.; Rienstra, C. M.
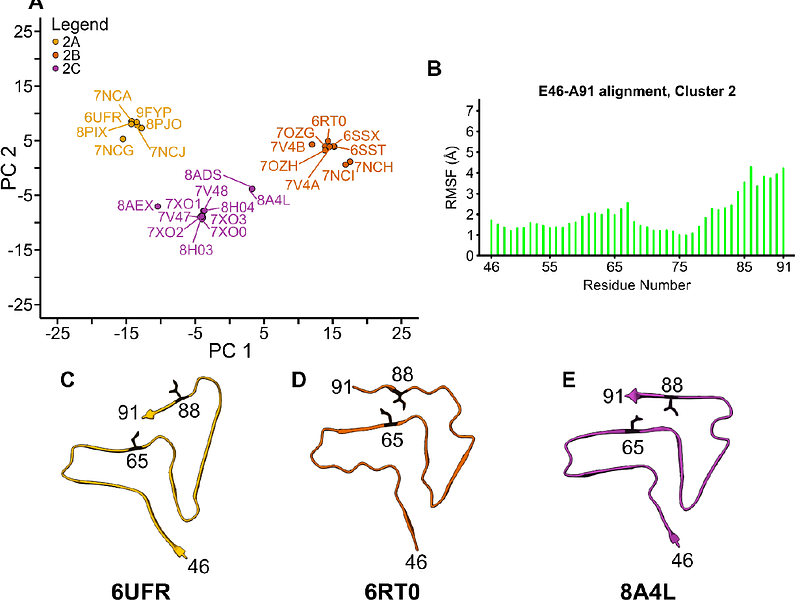
AbstractThe accumulation of Alpha-synuclein (Asyn) fibrils is the defining pathologic feature in Parkinson Disease (PD), Lewy Body Dementia (LBD), and Multiple System Atrophy (MSA). As such, the process of Asyn fibril formation has been an important research area and fibrils themselves have become attractive targets for disease diagnosis and therapeutic intervention. Due to the presence of mixed populations of fibrillar proteins associated with neurodegenerative diseases in brain tissue, high-resolution structures of Asyn fibrils are essential for the design of high-specificity imaging and therapeutic agents. Approximately one hundred high-resolution solid-state NMR (SSNMR) spectroscopy and cryo-electron microscopy (cryo-EM) structures of Asyn fibrils have been deposited to the Protein Databank (PDB); intriguingly there is significant polymorphism among them. Understanding the molecular makeup and characteristic features of each structural polymorph can determine conserved structural motifs which can be used as templates to design ligands with high specificity for clinical use. Utilizing standard alignment tools and density-based clustering approaches, we objectively classify fibril structures by tertiary structure type. We find that 81% of the structures cluster into two polymorph classes. Within each class, additional subtle variations are observed which position sidechains in specific, conserved orientations, well poised as druggable targets. Furthermore, we find that the conserved structural motifs associated with each class are found in all but one published Asyn fibril structure. We consider these classifications and conserved motifs in the context of disease-relevant fibril structures and offer a perspective on the utility of in vitro fibrils as substrates for drug development and models for disease pathogenesis.