A systematic approach to analyze T cell migration: application to mouse melanoma tumors
A systematic approach to analyze T cell migration: application to mouse melanoma tumors
Memmos, N.; Mitchell, J. S.; Fife, B. T.; Masopust, D.; Odde, D. J.
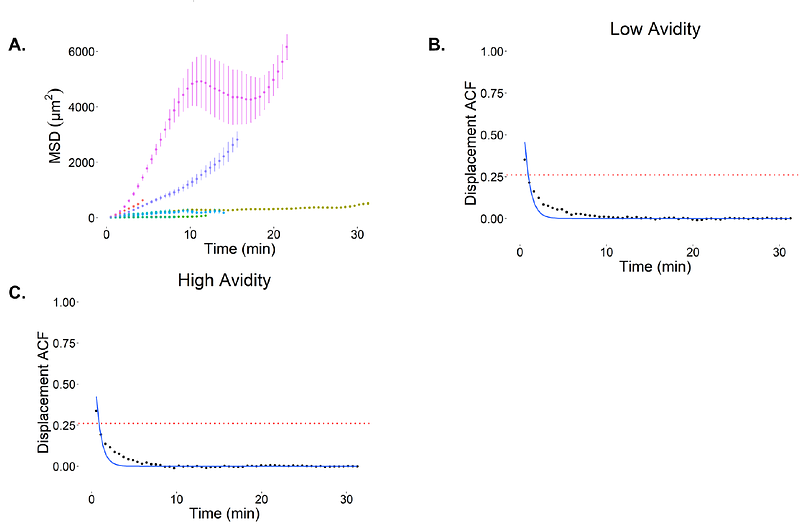
AbstractT cells must assess and choose between surveilling large areas, but also engage efficiently with the target cells. This process is translated into variations in speed and turning angle of T cells. In this study, we propose a generalized algorithm to analyze cell migration data with focus on CD8+ T cells, using clustering technique to identify the number of different migration states and Hidden Markov Model to capture the dynamical switching between them. The algorithm only requires a set of position observations in a series of times, independent of other factors. While this study focuses on CD8+ T cell migration, this approach can potentially be used broadly to study the migration of other cell types as well. For the current analysis, low and high avidity T cells in melanoma tumors were tracked ex vivo using two-photon microscopy. Our findings suggest that CD8+ T cells follow a two-state migration dynamic, with one state being faster, while the other slower and more localized. Moreover, we established a statistical methodology to analyze T cell migration to assess whether there is true variability in cell speeds as distinguished from stochastic fluctuations about a single speed, and it can be applied across different experimental platforms.