A single cell ATAC-seq atlas uncovers dynamic changes in chromatin accessibility during cell fate specification at the neural plate border
A single cell ATAC-seq atlas uncovers dynamic changes in chromatin accessibility during cell fate specification at the neural plate border
Harmrud, E.; Leese, J.; Thiery, A. P.; Buzzi, A. L.; Vigilante, A.; Briscoe, J.; Streit, A.
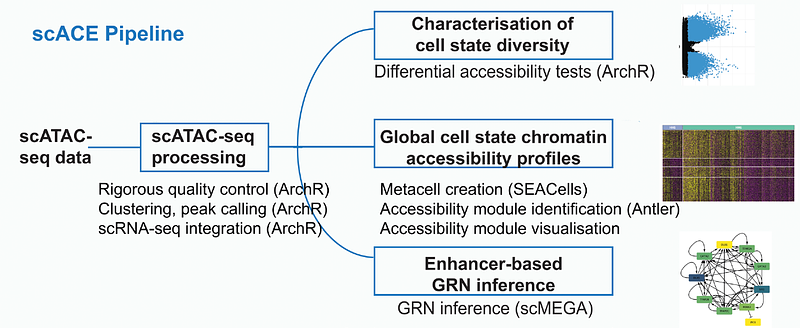
AbstractDuring development, dynamic changes in gene expression and chromatin architecture drive the transition from progenitors to specialised cell types. Here we use single cell ATAC sequencing (scATAC-seq) to investigate changes in chromatin accessibility as neural plate border cells segregate into neural, neural crest and placode cells. We developed a Nextflow pipeline, single cell Advanced Chromatin Exploration (scACE), which integrates scATAC-seq and scRNA-seq data to identify cell state specific accessibility profiles and groups of chromatin regions with coordinated dynamic behaviour, termed accessibility modules (AMs). We find that progenitors are characterised by broadly open chromatin, reflecting their broad potential to generate any ectodermal derivative. As development proceeds, cell-type-specific chromatin signatures are established. Inferring an enhancer-centric gene regulatory network, we predict Foxk2 as new regulator for placode specification and verify this prediction experimentally. Foxk2 target enhancers are open in placodal, but not any other ectodermal cells. This finding suggests that on a regulatory level, cells can use different strategies to control fate choice: differential accessibility of enhancers and broad accessibility controlled by differentially expressed transcription factors.