A functionally validated TCR-pMHC database for TCR specificity model development
A functionally validated TCR-pMHC database for TCR specificity model development
Messemaker, M.; Kwee, B. P. Y.; Moravec, Z.; Alvarez-Salmoral, D.; Urbanus, J.; de Paauw, S.; Geerligs, J.; Voogd, R.; Morris, B.; Guislain, A.; Mussmann, M.; Winkler, Y.; Steinmetz, M.; Iras, M.; Marcus, E.; Teuwen, J.; Perrakis, A.; Beijersbergen, R. L.; Scheper, W.; Schumacher, T.
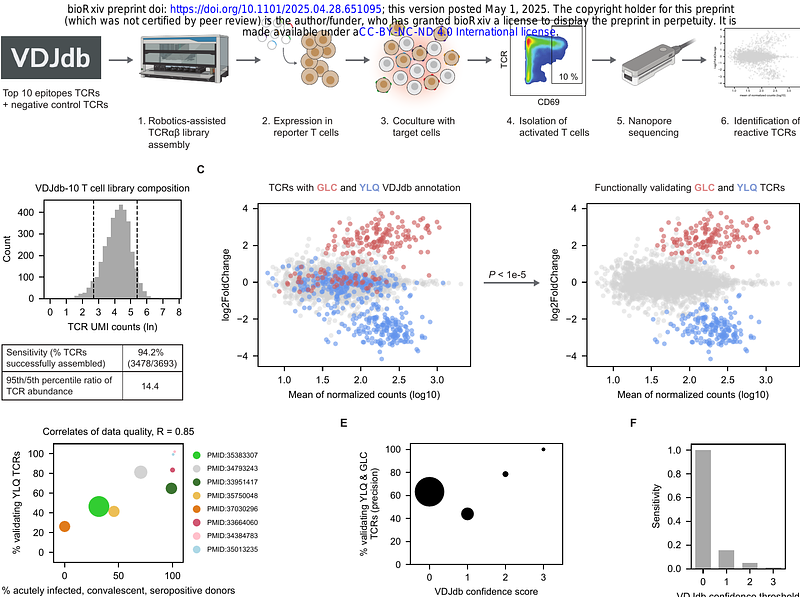
AbstractAccurate prediction of TCR specificity forms a holy grail in immunology and large language models and computational structure predictions provide a path to achieve this. Importantly, current TCR-pMHC prediction models have been trained and evaluated using historical data of unknown quality. Here, we develop and utilize a high-throughput synthetic platform for TCR assembly and evaluation to assess a large fraction of VDJdb-deposited TCR-pMHC entries using a standardized readout of TCR function. Strikingly, this analysis demonstrates that claimed TCR reactivity is only confirmed for 50% of evaluated entries. Intriguingly, the use of TCRbridge to analyze AlphaFold3 confidence metrics reveals a substantial performance in distinguishing functionally validating and non-validating TCRs even though AlphaFold3 was not trained on this task, demonstrating the utility of the validated VDJdb (TCRvdb) database that we generated. We provide TCRvdb as a resource to the community to support training and evaluation of improved predictive TCR specificity models.